Schedules
Amorphic Schedules automate processes and jobs in Amorphic. Users can configure custom schedules based on specific requirements.
Creating a Schedule
To create a schedule:
- Navigate to the Resource Details page.
- Click on the Schedules tab.
- Select
Add New Scheduleand provide the required details

Schedule configuration
| Type | Description |
|---|---|
| Schedule Name | A unique name that identifies the schedule's specific purpose. |
| Job Type | Users can select a job type from the dropdown list (details provided in the Job Type table below). |
| Schedule Type | Two types of schedules are available:
|
| Schedule Expression | Required for time-based schedules. Supports rate or cron expressions (e.g., Every 15 minutes, Daily, cron(0 12 * * ? *), etc.). |
Job Types
| Job Type | Description |
|---|---|
| ETL Job | Schedules an ETL job. |
| JDBC CDC | Synchronizes data between a data warehouse and S3 for Dataflows with Change Data Capture (CDC) process type. Only tasks with "SyncToS3" set to "yes" are visible for scheduling. |
| Data Ingestion | Schedules a data ingestion job for JDBC, S3, and external API data sources. |
| JDBC FullLoad | Schedules a JDBC Bulk Data Load full-load task. |
| DataPipelines | Schedules a DataPipeline execution. |
| Data Quality Checks | Schedules a data quality check for a dataset. |
| HCLS-Store | Schedules an import job for HealthLake Store, Omics Storage: Sequence Store, Omics Analytics: Variant Store, Annotation Store, and HealthImaging Store. |
| Health Image Data Conversion | Schedules a job which converts DICOM files in a dataset to NDJSON format and stores them in a different dataset. |
| Export to S3 | Schedules export to s3 operation for a dynamoDB Dataset . |
Supported Schedules Types by Resource
| Resource Type | Schedule Job Type |
|---|---|
| Datasets | Data Ingestion, Data Quality Checks, Export to S3, Health Image Data Conversion |
| Datasources | JDBC CDC, JDBC FullLoad |
| DataPipelines | DataPipelines |
| HCLS omics analytics | HCLS-Store |
| HCLS omics storage | HCLS-Store |
| HCLS healthlake | HCLS-Store |
| HCLS health imaging | HCLS-Store |
| Jobs | ETL Job |
If the schedule job type is 'Data Ingestion' and the dataset is of type 'reload', the execution will automatically reload the data.
-
Data Ingestion
Used to schedule a data ingestion job for supported data sources.
Supported Arguments
-
For JDBC Datasource Schedules
- NumberOfWorkers : Specifies the number of worker nodes allocated for the Glue job. (Valid range: 2–100).
- WorkerType: Specifies the worker type (computing resources) to use for the jobs. The worker type determines the amount of memory, CPU, and overall processing power allocated to each worker. Allowed values are
Standard, G.1X, G.2Xonly. - query : Allows users to specify a SQL SELECT query and ingest the data from source database retrieved by that SQL command.
- prepareQuery : Specifies a prefix that will form the final SQL query together with
queryargument. This argument offers a way to run such complex queries. Read here for more information.
-
For S3 and Ext-API Datasource Schedules
- MaxTimeOut : Overrides the default timeout setting of the datasource for the specific schedule (Valid range: 1–2880).
- MaxCapacity : Defines the number of AWS Glue data processing units (DPUs) that can be allocated when the job runs. (Allowed values: 1, 0.0625).
- FileConcurrency : Applicable to S3 data sources, determining the number of parallel file uploads.
-
-
Health Image Data Conversion
This schedule type converts DICOM files in a dataset to NDJSON format in order to upload it to Healthlake store, which only supports NDJSON file formats while importing data.
- Input dataset for these jobs must contain DICOM files.
- User have to specify output dataset id in arguments with key
outputDatasetIdand its value should be id of a valid dataset with Target Location as S3 and file type as others. - Converted NDJSON files will be stored into the specified output dataset.
- An optional argument
selectFilescan have the following values:- latest (default) – Selects only files uploaded after the last job run.
- all – Selects all files for conversion.
Schedule Details
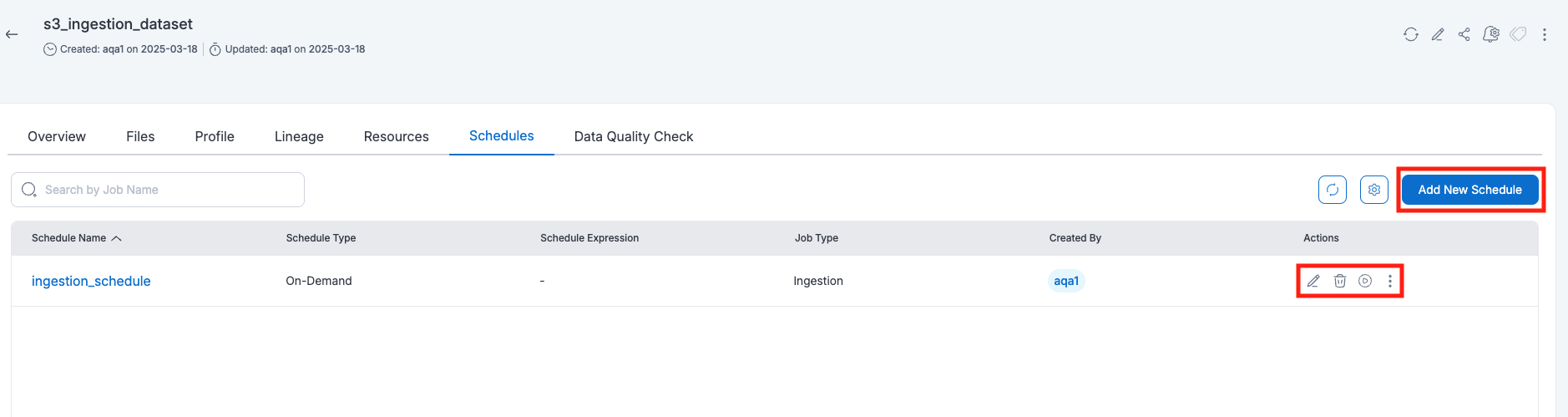
After creating a schedule, it will be listed on the Schedules page for the resource. Users can perform various actions, such as running, disabling, enabling, editing, cloning, or deleting the schedule.
Running a Schedule

Users can run a schedule by clicking the Run Schedule button on the schedule details page. To check the execution details, users can click on the schedule details, which provide information on whether the job is running, completed successfully, or failed.

- Schedule execution will fail if the related S3 datasource is using any of Amorphic S3 buckets as source. For ex:
<projectshortname-region-accountid-env-dlz> - For Data Ingestion Schedules, the following arguments can be provided during schedule runs:
- MaxTimeOut: This argument allows users to override the timeout setting of the datasource for the specific run. It accepts values from 1 to 2880.
- FileConcurrency: This argument enables users to configure the number of parallel file ingestion that occur for S3 datasource. It accepts values from 1 to 100 and has a default value of 20.
Event Trigger Schedules
Event Trigger schedules automatically execute when specific events occur in your system. For example, you can configure a schedule to run automatically when new files are uploaded to a dataset. This automation is particularly useful for triggering Targets such as data pipelines or ETL jobs in response to changes in system.
Creating an Event Trigger Schedule
To create an event trigger schedule:
- Navigate to the Resource Details page.
- Click on the Schedules tab.
- Select
Create Scheduleand provide the required details. - Select
Event Triggeras the schedule type. - Select the event type and specify the Targets.
- Click
Createto save the schedule.
Event Types
The following Event Types are currently supported for Event Trigger Schedules:
- File Upload: Invokes targets for each file uploaded to dataset
- Ingestion: Invokes targets for each successfull ingestion job completion
Once created, you can manage the event trigger schedule by:
- Enabling or Disabling the schedule
- Editing the description and target configurations
- Deleting the schedule when no longer needed
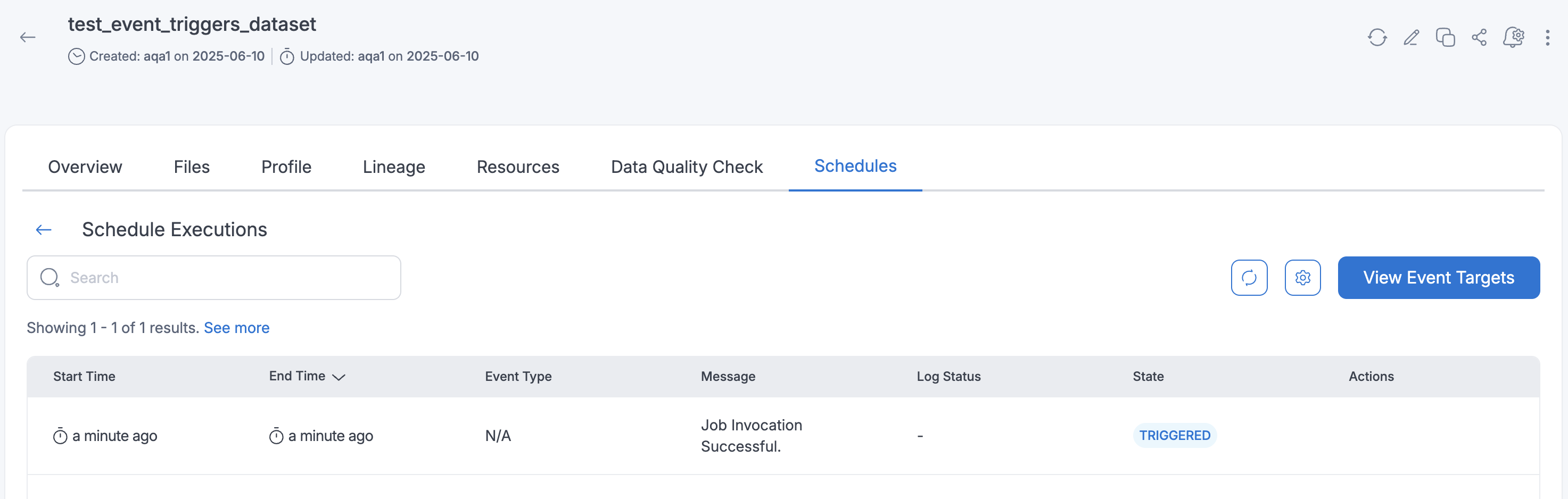
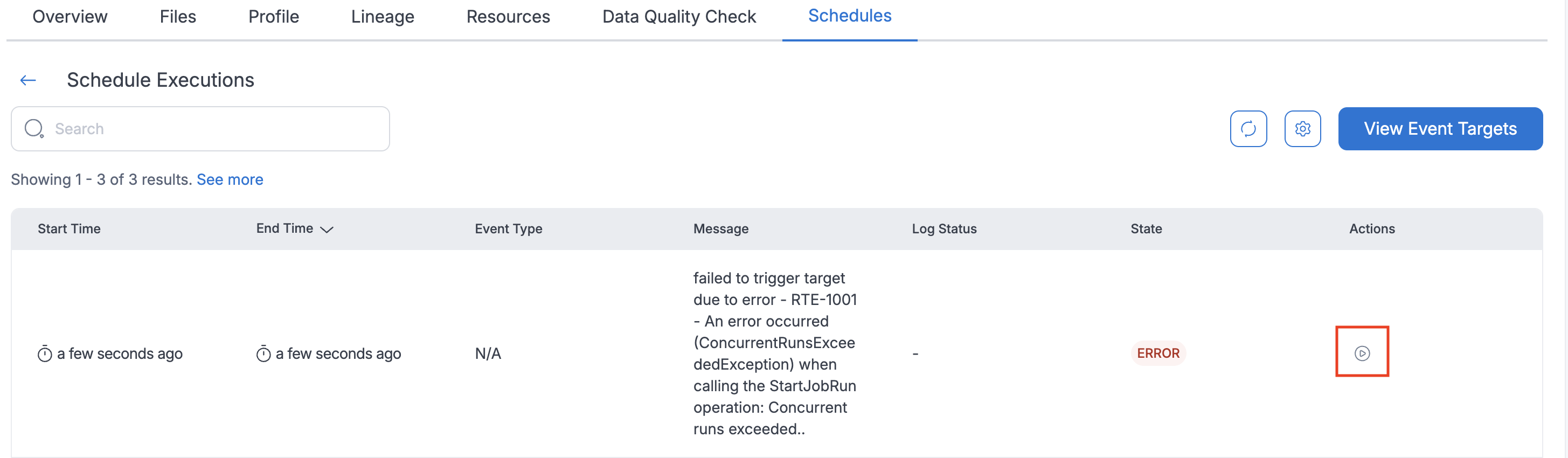
Rerunning Failed Event Trigger Executions
If an event trigger schedule execution fails, you can manually restart it:
- Navigate to the schedule executions page.
- Click the
Re-Triggerbutton for the failed execution. - The system will re-trigger the failed execution.
- Currently available only for Dataset resources
- Supported event type: File uploads to datasets and Ingestion job completion
- Supported target types: Data pipelines and ETL jobs
- Event triggers are not instantaneous and may experience delays based on system load
Schedule use case
When the schedule execution is completed, an email notification will be sent out, based on the notification setting and schedule execution status. Users can also view the execution logs of each schedule run, which includes Output Logs, Output Logs (Full), and Error Logs.
For example, if User needs to create a schedule that runs an ETL job and sends out important emails every 4 hours, user can create a DataPipeline with an ETL Job Node followed by a Mail Node. This workflow can then be scheduled to run every 4 hours, every day.
to see how to create schedules on resource, check How to create schedules